Health Knowledge Graph Extraction (HERON)
2022 — Principal Investigator
This internally funded research effort sought to formalized unstructured information: transforming dense, human-interpretable knowledge (e.g., academic articles) into a machine- readable form (knowledge graphs). Specifically, HERON focused on named entity recognition (NER) to identify entities of interest, relation extraction (RE) to determine whether and how those entities relate, and entity linking to associate those entities with existing knowledge bases.
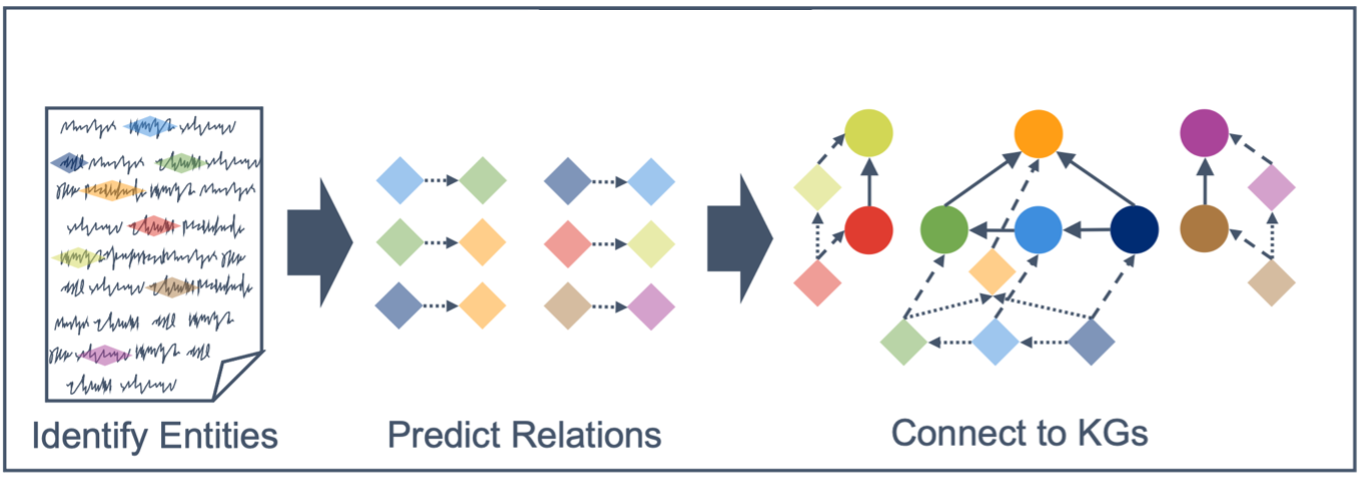
As part of this work, we developed CNN-based methods for NER and RE in the biomedical domain (e.g., PubMed articles), and showed improved performance over fine-tuned baselines (e.g., SciBERT). We also explored methods for leveraging transformer-based question- answering models to weakly annotate publicly available NER data (e.g., MedMentions) with relation labels for use in RE training with promising results. A poster describing this work was presented at the National Library of Medicine Curation at Scale workshop (March 2022).
